Editor's Note: This article first appeared in the March 23 issue of The Daily Texan.
The University is partnering with the Texas Department of State Health Services to analyze COVID-19 samples from across Texas, allowing them to detect potential variants.
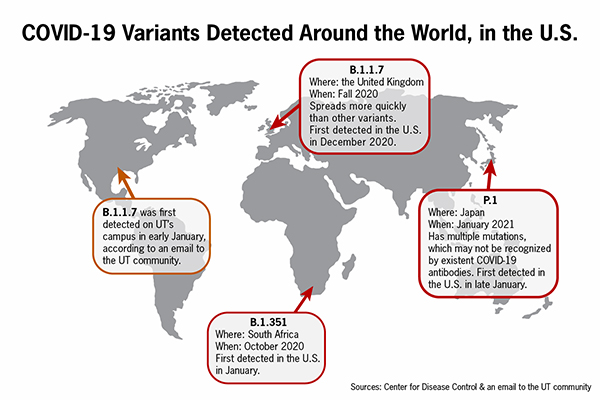
UT’s Center for Biomedical Research Support, which provides technological support to research projects, currently uses samples from Proactive Community Testing to detect variants in UT’s population. Both the United Kingdom variant B.1.1.7 and the California variant B.1.429 have been detected in the UT community through proactive testing.
“Since the (Genomic Sequencing and Analysis Facility) here at UT has sequencing machines designed for high capacity sequencing, we should be able to increase the efficiency of SARS-CoV-2 sequencing for both DSHS and UT,” said Dennis Wylie, research scientist for computational biology and bioinformatics.
The University’s sequencing machine can test up to 700 COVID-19 samples at a time, but the University only receives around 200 samples a day, said Rachel Lee, microbiological sciences branch manager for the Department of State Health Services. The department receives more samples than the University but has smaller sequencing machines, Lee said.
“We don't have to run five different machines for 500 samples, we can just use UT’s one,” Lee said. “We give them 500 samples, they have 200 samples. They don't have to waste their reagents. It all fits in one plate — it’s perfect for everyone.”
The Center for Biomedical Research Support collects positive COVID-19 samples from the University’s Proactive Community Testing program and sequences them through a machine, which tests the sample’s viral genome, said Andreas Matouschek, director for the Center for Biomedical Research Support. The viral genome is the virus’ blueprint and contains the information to make more COVID-19 cells, Matouschek said.
After sequencing, the center assembles the information from sequencing and analyzes it to detect mutations in the virus, which eventually lead to variants, Matouschek said.
“Mutations in the virus are not uncommon so you'll see a blip here and you'll see a blip there, and it's hard to know what it means,” Matouschek said. “But over time, people have discovered that a combination of these makes a new strain of virus.”
Lee said the department approached the University for a partnership to better understand COVID-19 variants across the state.
“Once we get our sequencing results, we have so many samples with so many different COVID strains,” Lee said. “We talked about how we can work together to expand the sequencing capacity for both laboratories and to be able … to understand the prevalence and distribution of different variants in the state of Texas.”
DSHS receives samples from the state’s Laboratory Response Network and commercial laboratories, which includes San Antonio, El Paso, Dallas, Harlingen and Houston, Lee said.
“When they receive samples, and the COVID samples test positive and meet the criteria, then they'll send it to us,” Lee said. “If anybody suspects UK variants or other variants of concern, they send it to us for sequencing.”
DSHS is drafting contracts with the University in order to begin sequencing and analyzing efforts, Lee said. She hopes to have an “in-house pipeline” between the University and DSHS within the next few weeks.