In response to the dilemma of bacterial antibiotic resistance, a research laboratory at UT-Austin has developed a novel method of screening named SLAY, for Surface Localized Antimicrobial Display.
“Antibiotic-resistant bacterial infections are a growing medical problem,” said Bryan Davies, an assistant professor in molecular biosciences.
As a result, the supply of available effective antibiotics is dwindling, said Ashley Tucker, a postdoctoral fellow at UT and member of the Davies Lab. Pharmaceutical companies have resorted to deriving already existing antibiotics that have been around for the last 40 years.
“The bacteria will always win, because they are incredible at adapting to challenges, but we can get one step ahead if we can more quickly discover new scaffolds for antibiotics,” Tucker said.
Antibiotic resistance arises when bacteria mutate their own genomes or acquire DNA that encodes for antibiotic resistance, Davies explained. Because of this, it can be very challenging to identify new antibiotic leads, but SLAY provides a new way to do so.
“You need to screen drugs to find a lead antibiotic,” Davies said. “SLAY allows us to screen large numbers of peptides at once, (and the) more peptides screened, the (better) chance of finding a good lead antibiotic.”
Essentially, SLAY is a method that puts bacteria to work for the researchers, Tucker said.
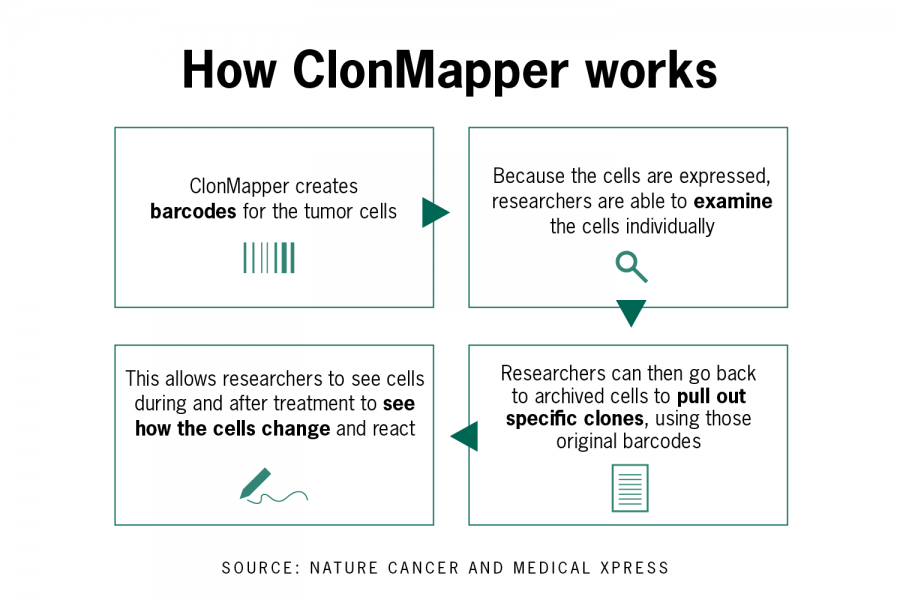
SLAY allows bacteria to make their own unique genetically encoded peptide — a simpler protein structure — and tether the peptide to its surface. If the peptide kills the bacteria, there exists potential antimicrobial properties and the amino acid sequence of the peptide can then be determined using next-generation sequencing.
“The beauty of SLAY is the sheer chemical diversity that you are able to sample when you screen upwards of a million compounds at once,” Tucker said. “We have a real potential to find antibiotic chemistry that nature hasn’t yet tapped into.”
SLAY is faster, cheaper and more effective than any current drug-screening platform on the market, and it has received an overwhelmingly positive response from academia, industry and military collaborators, Tucker said. This project was supported by the National Institutes of Health, Sanofi, the Welch Foundation, the Defense Advanced Research Projects Agency and the U.S. Army Research Office.
“We ultimately hope to see multiple groups using our SLAY technology to fight the problem of antimicrobial resistance because having that many groups working toward a single goal together will give the world a better chance at finding antibiotics that really work,” Tucker said.
And SLAY’s novelty doesn’t just lie in its efficacy and potential — there’s also a double meaning to the method’s name.
“The timing of coming up with an acronym happened to coincide with Beyonce’s viral pregnancy announcement — and that’s what sparked the idea of blending science with pop culture — to get people really excited about our technology,” Tucker said. “Beyonce is a native Texan, a force in the music industry, and has really popularized the term ‘slay,’ and so the concept just worked. You can also think about ‘slay’ in terms of killing, which is exactly the intention of the technology – to find peptides that slay the bacteria.”
The battle isn’t over yet, though.
“I hope SLAY will spark something in the antibiotic research community to think outside of the box,” Tucker said.